Polysome profiling
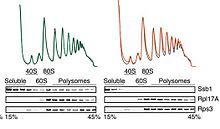
Polysome profiling is a technique in molecular biology that is used to study the association of mRNAs with ribosomes. It is important to note that this technique is different from ribosome profiling. Both techniques have been reviewed[1] and both are used in analysis of the translatome, but the data they generate are at very different levels of specificity. When employed by experts, the technique is remarkably reproducible: the 3 profiles in the first image are from 3 different experiments.[2]
The procedure[edit]
The procedure begins by making a cell lysate of the cells of interest. This lysate contains polysomes, monosomes (composed of one ribosome residing on an mRNA), the small (40S in eukaryotes) and large (60S in eukaryotes) ribosomal subunits, "free" mRNA and a host of other soluble cellular components.
The procedure continues by making a continuous sucrose gradient of continuously variable density in a centrifuge tube. At the concentrations used (15-45% in the example), sucrose does not disrupt the association of ribosomes and mRNA. The 15% portion of the gradient is at the top of the tube, while the 45% portion is at the bottom because of their different density.
A specific amount (as measured by optical density) of the lysate is then layered gently on top of the gradient in the tube. The lysate, even though it contains a large amount of soluble material, is much less dense than 15% sucrose, and so it can be kept as a separate layer at the top of the tube if this is done gently.
In order to separate the components of the lysate, the preparation is subjected to centrifugation. This accelerates the components of the lysate with many times the force of gravity and thus propels them through the gradient based upon how "big" the individual components are. The small (40S) subunits travel less far into the gradient than the large (60S) subunits. The 80S ribosomes on an mRNA travel further (note that the contribution of the size of the mRNA to the distance traveled is not significant). Polysomes composed of 2 ribosomes travel further, polysomes with 3 ribosomes travel further still, and on and on. The "size" of the components is designated by S, the svedberg unit. Note that one S = 10−13 seconds, and that the concept of "big" is actually an oversimplification.

After centrifugation, the contents of the tube are collected as fractions from the top (smaller, slower traveling) to bottom (bigger, faster traveling) and the optical density of the fractions is determined. The first fractions removed have a large amount of relatively small molecules, such as tRNAs, individual proteins, etc.
Applications[edit]
It is possible to use this technique to study the overall degree of translation in cells (for examples[3][4][5]), but it can be used much more specifically to study individual proteins and their mRNAs. As an example shown in the lower portion of the figure, a protein that composes part of the small subunit can first be detected in the 40S fraction, then nearly disappears from the 60S fraction (the separations on these gradients are not absolute), then reappears in the 80S and polysome fractions. This indicates that there is at most very little of the protein found in the cell that is not part of the small subunit. In contrast, in the upper row of the immunoblot figure, a soluble protein appears in the soluble fractions and associated with ribosomes and polysomes. The particular protein is a chaperone protein, which (in brief) helps to fold the nascent peptide as it is being extruded from the ribosome. As other work
in the paper showed, there is a direct association of the chaperone with the ribosome.[2]
The technique can also be used to study the degree of translation of a particular mRNA[6] In these experiments, 5' and 3' sequences of an mRNA were investigated for their effects on amount of mRNA produced and how well the mRNAs were translated. As shown, not all mRNA isoforms are translated with the same efficiency even though their coding sequences are the same.[6]
References[edit]
- ^ Piccirillo, CA; et al. (2014). "Translational control of immune responses: from transcripts to translatomes". Nature Immunology. 15 (6): 503–511. doi:10.1038/ni.2891. PMID 24840981. S2CID 6269940.
- ^ a b Hanebuth, MA; et al. (2016). "Multivalent contacts of the Hsp70 Ssb contribute to its architecture on ribosomes and nascent chain interaction". Nature Communications. 7: 13695. Bibcode:2016NatCo...713695H. doi:10.1038/ncomms13695. PMC 5150220. PMID 27917864.
- ^ Lin, CJ; et al. (2010). "The antidepressant sertraline inhibits translation initiation by curtailing mammalian target of rapamycin signaling" (PDF). Cancer Research. 70 (8): 3199–3208. doi:10.1158/0008-5472.CAN-09-4072. PMID 20354178.
- ^ Coudert, L; et al. (2014). "Analysis of translation initiation during stress conditions by polysome profiling". Journal of Visualized Experiments (87). doi:10.3791/51164. PMC 4193336. PMID 24893838.
- ^ Molon, M; et al. (2016). "The rate of metabolism as a factor determining longevity of the Saccharomyces cerevisiae yeast". Age (Dordrecht, Netherlands). 38 (1): 11. doi:10.1007/s11357-015-9868-8. PMC 5005888. PMID 26783001.
- ^ a b Floor, SN; Doudna, JA (2016). "Tunable protein synthesis by transcript isoforms in human cells". eLife. 5. doi:10.7554/eLife.10921. PMC 4764583. PMID 26735365.
