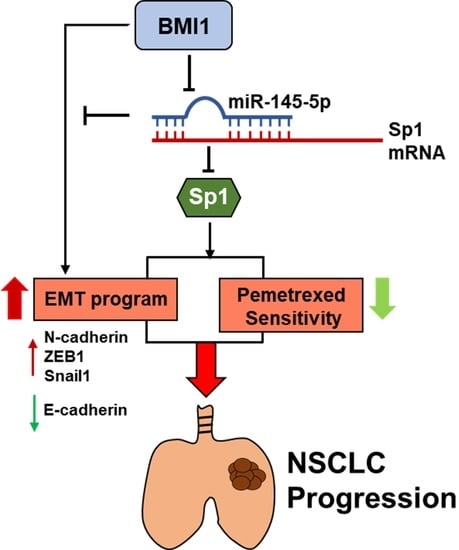
miR-145-5p Targets Sp1 in Non-Small Cell Lung Cancer Cells and Links to BMI1 Induced Pemetrexed Resistance and Epithelial–Mesenchymal Transition
Abstract
:1. Introduction
2. Results
2.1. The Expression Levels of miR-145-5p Change Pemetrexed Sensitivity of NSCLC Cells
2.2. Sp1 Is a Direct Target of miR-145-5p in NSCLC Cells
2.3. Overexpression of Sp1 Decreases Pemetrexed Sensitivity and Induction EMT in NSCLC Cells
2.4. miR-145-5p Suppresses BMI1 Induced Cell Proliferation and EMT in NSCLC Cells
2.5. BMI1 Induced Malignant Phenotypes in NSCLC Cells Can Be Reduced by Sp1 Inhibition
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Determination of Pemetrexed Sensitivity
4.3. Cell Transfection
4.4. Reverse Transcription Quantitative PCR (RT-qPCR)
4.5. Western Blotting
4.6. Luciferase Reporter Assay
4.7. Wound-Healing Assay
4.8. Clonogenic Assay
4.9. Analysis of TCGA Data
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oser, M.G.; Niederst, M.J.; Sequist, L.V.; Engelman, J.A. Transformation from non-small-cell lung cancer to small-cell lung cancer: Molecular drivers and cells of origin. Lancet Oncol. 2015, 16, e165–e172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walser, T.; Cui, X.; Yanagawa, J.; Lee, J.M.; Heinrich, E.; Lee, G.; Sharma, S.; Dubinett, S.M. Smoking and lung cancer: The role of inflammation. Proc. Am. Thorac. Soc. 2008, 5, 811–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Goldberg, M.S.; Villeneuve, P.J. A Systematic Review of Relation between Long-term Exposure to Ambient Air Pollution and Chronic Disease. Rev. Environ. Health 2008, 23, 243–297. [Google Scholar] [CrossRef] [PubMed]
- Longo, D.L.; Fauci, A.S.; Kasper, D.L.; Hauser, S.L.; Jameson, J.L.; Loscalzo, J. Harrison’s Principles of Internal Medicine; Mcgraw-Hill: New York, NY, USA, 2012; Volume 2012. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chang, Y.J.; Huang, J.Y.; Lin, C.H.; Wang, B.Y. Survival and Treatment of Lung Cancer in Taiwan between 2010 and 2016. J. Clin. Med. 2021, 10, 4675. [Google Scholar] [CrossRef]
- Herbst, R.S.; Heymach, J.V.; Lippman, S. Molecular origins of cancer. N. Engl. J. Med. 2008, 359, 1367–1380. [Google Scholar] [CrossRef] [Green Version]
- Tomasini, P.; Barlesi, F.; Mascaux, C.; Greillier, L. Pemetrexed for advanced stage nonsquamous non-small cell lung cancer: Latest evidence about its extended use and outcomes. Ther. Adv. Med. Oncol. 2016, 8, 198–208. [Google Scholar] [CrossRef] [Green Version]
- Chiu, L.; Hsin, I.; Yang, T.; Sung, W.; Chi, J.; Chang, J.; Ko, J.; Sheu, G. The ERK–ZEB1 pathway mediates epithelial-mesenchymal transition in pemetrexed resistant lung cancer cells with suppression by vinca alkaloids. Oncogene 2017, 36, 242–253. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, R.; Pan, Y.; Sun, Y.; Zhang, J.; Chen, H. The pemetrexed-containing treatments in the non-small cell lung cancer, is-/low thymidylate synthase expression better than+/high thymidylate synthase expression: A meta-analysis. BMC Cancer 2014, 14, 205. [Google Scholar]
- Bukhari, A.A.; Goudar, R.K. Thymidylate synthase as a predictive biomarker for pemetrexed response in NSCLC. Lung Cancer Int. 2013, 2013, 436409. [Google Scholar] [CrossRef] [Green Version]
- He, Y.-W.; Zhao, M.-L.; Yang, X.-Y.; Zeng, J.; Deng, Q.-H.; He, J.-X. Prognostic value of ERCC1, RRM1, and TS proteins in patients with resected non-small cell lung cancer. Cancer Chemother. Pharmacol. 2015, 75, 861–867. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.T.; Chien, P.J.; Chen, S.H.; Sheu, G.T.; Jan, M.S.; Wang, B.Y.; Chang, W.W. BMI1-Mediated Pemetrexed Resistance in Non-Small Cell Lung Cancer Cells Is Associated with Increased SP1 Activation and Cancer Stemness. Cancers 2020, 12, 2069. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.Y.; Wang, R.; Chen, L.B. Micro RNA-145: A potent tumour suppressor that regulates multiple cellular pathways. J. Cell. Mol. Med. 2014, 18, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Mataki, H.; Seki, N.; Mizuno, K.; Nohata, N.; Kamikawaji, K.; Kumamoto, T.; Koshizuka, K.; Goto, Y.; Inoue, H. Dual-strand tumor-suppressor microRNA-145 (miR-145–5p and miR-145–3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016, 7, 72084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, T.-Q.; Xie, Z.-C.; Tang, R.-X.; Zhang, T.-T.; Li, D.-Y.; Li, Z.-Y.; Chen, G. Clinical value of miR-145–5p in NSCLC and potential molecular mechanism exploration: A retrospective study based on GEO, qRT-PCR, and TCGA data. Tumor Biol. 2017, 39, 1010428317691683. [Google Scholar] [CrossRef] [Green Version]
- Mei, L.-L.; Wang, W.-J.; Qiu, Y.-T.; Xie, X.-F.; Bai, J.; Shi, Z.-Z. miR-145–5p suppresses tumor cell migration, invasion and epithelial to mesenchymal transition by regulating the Sp1/NF-κB signaling pathway in esophageal squamous cell carcinoma. Int. J. Mol. Sci. 2017, 18, 1833. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Jiang, M.; Liu, Q.; Han, Z.; Zhao, Y.; Ji, S. miR-145–5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol. Lett. 2018, 16, 6355–6360. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zhang, J.; Tong, Y.; Li, J.; Liu, B. miR-145–5p restrained cell growth, invasion, migration and tumorigenesis via modulating RHBDD1 in colorectal cancer via the EGFR-associated signaling pathway. Int. J. Biochem. Cell Biol. 2019, 117, 105641. [Google Scholar] [CrossRef]
- Chang, Y.; Yan, W.; Sun, C.; Liu, Q.; Wang, J.; Wang, M. miR-145-5p inhibits epithelial-mesenchymal transition via the JNK signaling pathway by targeting MAP3K1 in non-small cell lung cancer cells. Oncol. Lett. 2017, 14, 6923–6928. [Google Scholar] [CrossRef]
- Zheng, F.; Xu, R. CircPVT1 contributes to chemotherapy resistance of lung adenocarcinoma through miR-145–5p/ABCC1 axis. Biomed. Pharmacother. 2020, 124, 109828. [Google Scholar] [CrossRef]
- Qiu, T.; Zhou, X.; Wang, J.; Du, Y.; Xu, J.; Huang, Z.; Zhu, W.; Shu, Y.; Liu, P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014, 588, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.K.; Lee, Y.S.; Kim, H.S.; Dong, S.M.; Park, J.S.; Yoon, D.S. Specific protein 1(SP1) regulates the epithelial-mesenchymal transition via lysyl oxidase-like 2(LOXL2) in pancreatic ductal adenocarcinoma. Sci. Rep. 2019, 9, 5933–5944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Bracken, C.P.; Szubert, J.M.; Goodall, G.J. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS ONE 2013, 8, e55214–e55220. [Google Scholar] [CrossRef] [Green Version]
- Takezawa, K.; Okamoto, I.; Okamoto, W.; Takeda, M.; Sakai, K.; Tsukioka, S.; Kuwata, K.; Yamaguchi, H.; Nishio, K.; Nakagawa, K. Thymidylate synthase as a determinant of pemetrexed sensitivity in non-small cell lung cancer. Br. J. Cancer 2011, 104, 1594–1601. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.H.; Ahn, J.S.; Lee, J.; Kim, K.H.; Park, Y.H.; Han, J.; Ahn, M.J.; Park, K. The efficacy of pemetrexed as a third- or fourth-line therapy and the significance of thymidylate synthase expression in patients with advanced non-small cell lung cancer. Lung Cancer 2010, 69, 323–329. [Google Scholar] [CrossRef]
- Liang, S.Q.; Marti, T.M.; Dorn, P.; Froment, L.; Hall, S.R.; Berezowska, S.; Kocher, G.; Schmid, R.A.; Peng, R.W. Blocking the epithelial-to-mesenchymal transition pathway abrogates resistance to anti-folate chemotherapy in lung cancer. Cell Death Dis. 2015, 6, e1824. [Google Scholar] [CrossRef] [Green Version]
- Koren, A.; Rijavec, M.; Sodja, E.; Kern, I.; Sadikov, A.; Kovac, V.; Korosec, P.; Cufer, T. High BMI1 mRNA expression in peripheral whole blood is associated with favorable prognosis in advanced non-small cell lung cancer patients. Oncotarget 2017, 8, 25384–25394. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, T.; Sun, W.; Liu, C.; Fang, X. Bmi-1 overexpression as an efficient prognostic marker in patients with nonsmall cell lung cancer. Medicine 2017, 96, e7346. [Google Scholar] [CrossRef]
- Lopez-Arribillaga, E.; Rodilla, V.; Pellegrinet, L.; Guiu, J.; Iglesias, M.; Roman, A.C.; Gutarra, S.; Gonzalez, S.; Munoz-Canoves, P.; Fernandez-Salguero, P.; et al. Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development 2015, 142, 41–50. [Google Scholar] [CrossRef]
- Paranjape, A.N.; Balaji, S.A.; Mandal, T.; Krushik, E.V.; Nagaraj, P.; Mukherjee, G.; Rangarajan, A. Bmi1 regulates self-renewal and epithelial to mesenchymal transition in breast cancer cells through Nanog. BMC Cancer 2014, 14, 785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.; Mirshahidi, S.; Simental, A.; Lee, S.C.; de Andrade Filho, P.A.; Peterson, N.R.; Duerksen-Hughes, P.; Yuan, X. Cancer stem cell self-renewal as a therapeutic target in human oral cancer. Oncogene 2019, 38, 5440–5456. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Bharali, D.J.; Sudha, T.; Khedr, M.; Guest, I.; Sell, S.; Glinsky, G.V.; Mousa, S.A. Downregulation of Bmi1 in breast cancer stem cells suppresses tumor growth and proliferation. Oncotarget 2017, 8, 38731–38742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Lee, S.; Park, W.H.; Suh, D.H.; Kim, K.; Kim, Y.B.; No, J.H. Silencing Bmi1 expression suppresses cancer stemness and enhances chemosensitivity in endometrial cancer cells. Biomed. Pharmacother. 2018, 108, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.H.; Yang, S.F.; Li, P.Q. Human lung cancer cell line SPC-A1 contains cells with characteristics of cancer stem cells. Neoplasma 2012, 59, 685–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toden, S.; Tran, H.M.; Tovar-Camargo, O.A.; Okugawa, Y.; Goel, A. Epigallocatechin-3-gallate targets cancer stem-like cells and enhances 5-fluorouracil chemosensitivity in colorectal cancer. Oncotarget 2016, 7, 16158–16171. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.B.; Liu, G.H.; Zhang, H.; Xing, S.; Hu, L.J.; Zhao, W.F.; Xie, B.; Li, M.Z.; Zeng, B.H.; Li, Y.; et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 2013, 280, 2929–2944. [Google Scholar] [CrossRef]
- Zhang, J.P.; Zhang, H.; Wang, H.B.; Li, Y.X.; Liu, G.H.; Xing, S.; Li, M.Z.; Zeng, M.S. Down-regulation of Sp1 suppresses cell proliferation, clonogenicity and the expressions of stem cell markers in nasopharyngeal carcinoma. J. Transl. Med. 2014, 12, 222. [Google Scholar] [CrossRef] [Green Version]
- Polytarchou, C.; Iliopoulos, D.; Struhl, K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc. Natl. Acad. Sci. USA 2012, 109, 14470–14475. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, W.-W.; Wang, B.-Y.; Chen, S.-H.; Chien, P.-J.; Sheu, G.-T.; Lin, C.-H. miR-145-5p Targets Sp1 in Non-Small Cell Lung Cancer Cells and Links to BMI1 Induced Pemetrexed Resistance and Epithelial–Mesenchymal Transition. Int. J. Mol. Sci. 2022, 23, 15352. https://doi.org/10.3390/ijms232315352
Chang W-W, Wang B-Y, Chen S-H, Chien P-J, Sheu G-T, Lin C-H. miR-145-5p Targets Sp1 in Non-Small Cell Lung Cancer Cells and Links to BMI1 Induced Pemetrexed Resistance and Epithelial–Mesenchymal Transition. International Journal of Molecular Sciences. 2022; 23(23):15352. https://doi.org/10.3390/ijms232315352
Chicago/Turabian StyleChang, Wen-Wei, Bing-Yen Wang, Shih-Hong Chen, Peng-Ju Chien, Gwo-Tarng Sheu, and Ching-Hsiung Lin. 2022. "miR-145-5p Targets Sp1 in Non-Small Cell Lung Cancer Cells and Links to BMI1 Induced Pemetrexed Resistance and Epithelial–Mesenchymal Transition" International Journal of Molecular Sciences 23, no. 23: 15352. https://doi.org/10.3390/ijms232315352