Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process
Abstract
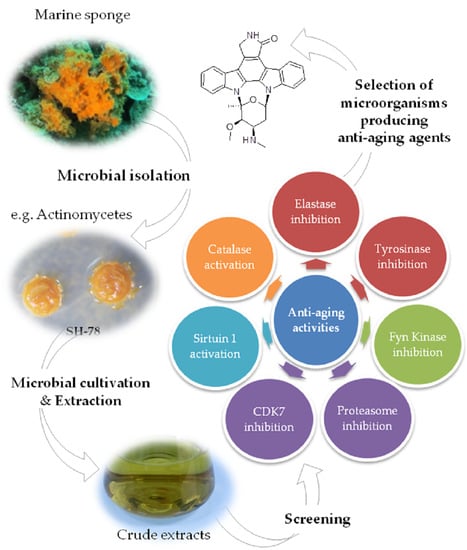
:1. Introduction
2. Results and Discussion
2.1. Composition of the Prokaryotic Community of Scopalina hapalia
2.2. Isolation and Identification of Microorganisms Associated with Scopalina hapalia
2.2.1. Identification of Culturable Actinomycetes Associated with Scopalina hapalia
2.2.2. Identification of Bacillales Strains Isolated from Scopalina hapalia
2.2.3. Identification of Filamentous Fungi
2.3. Biological Assays
2.3.1. Anti-Elastase Activities
2.3.2. Anti-Tyrosinase Activities
2.3.3. Catalase Activities
2.3.4. Sirtuin 1 Activities
2.3.5. Anti-CDK7 Activities
2.3.6. Anti-Fyn Kinase Activities
2.3.7. Anti-Proteasome Activities
3. Materials and Methods
3.1. Sponge Collection
3.2. Targeted 16S rRNA Gene Sequencing / Next Generation Sequencing
3.3. Alpha Diversity Analysis
3.4. Microbial Isolation
3.5. Molecular Identification
3.6. Extracts Preparation
3.7. Bioassays
3.7.1. Elastase Activity Assay
3.7.2. Tyrosinase Activity Assay
3.7.3. Catalase Activation Assay
3.7.4. Sirtuin 1 Activation Assay
3.7.5. CDK7 Inhibition Assay
3.7.6. FynB Inhibition Activity
3.7.7. Proteasome Inhibition Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Niccoli, T.; Partridge, L. Ageing as a Risk Factor for Disease. Curr. Biol. 2012, 22, R741–R752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, B.K.; Berger, S.L.; Brunet, A.; Campisi, J.; Cuervo, A.M.; Epel, E.S.; Franceschi, C.; Lithgow, G.J.; Morimoto, R.I.; Pessin, J.E.; et al. Geroscience: Linking aging to chronic disease. Cell 2014, 159, 709–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longo, V.D.; Antebi, A.; Bartke, A.; Barzilai, N.; Brown-Borg, H.M.; Caruso, C.; Curiel, T.J.; Cabo, R.; De Franceschi, C.; Gems, D.; et al. Interventions to slow aging in humans: Are we ready? Aging Cell 2015, 14, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span-from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Davinelli, S.; Willcox, D.C.; Scapagnini, G. Extending healthy ageing: Nutrient sensitive pathway and centenarian population. Immun. Ageing 2012, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Seals, D.R.; Justice, J.N.; LaRocca, T.J. Physiological geroscience: Targeting function to increase healthspan and achieve optimal longevity. J. Physiol. 2016, 594, 2001–2024. [Google Scholar] [CrossRef]
- Robert, L.; Jacob, M.P.; Frances, C.; Godeau, G.; Hornebeck, W. Interaction between elastin and elastases and its role in the aging of the arterial wall, skin and other connective tissues. A review. Mech. Ageing Dev. 1984, 28, 155–166. [Google Scholar] [CrossRef]
- Tsuji, N.; Moriwaki, S.; Suzuki, Y.; Takema, Y.; Imokawa, G. The role of elastases secreted by fibroblasts in wrinkle formation: Implication through selective inhibition of elastase activity. Photochem. Photobiol. 2001, 74, 283–290. [Google Scholar] [CrossRef]
- Rijken, F.; Kiekens, R.C.M.; Bruijnzeel, P.L.B. Skin-infiltrating neutrophils following exposure to solar-simulated radiation could play an important role in photoageing of human skin. Br. J. Dermatol. 2005, 152, 321–328. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V. Skin whitening agents: Medicinal chemistry perspective of tyrosinase inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 403–425. [Google Scholar] [CrossRef] [Green Version]
- Bae-Harboe, Y.-S.C.; Park, H.-Y. Tyrosinase: A Central Regulatory Protein for Cutaneous Pigmentation. J. Investig. Dermatol. 2012, 132, 2678–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hekimi, S.; Lapointe, J.; Wen, Y. Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 2011, 21, 569–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glorieux, C.; Calderon, P.B. Catalase, a remarkable enzyme: Targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 2017, 398, 1095–1108. [Google Scholar] [CrossRef] [Green Version]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Longo, V.D.; Kennedy, B.K. Sirtuins in Aging and Age-Related Disease. Cell 2006, 126, 257–268. [Google Scholar] [CrossRef] [Green Version]
- Bordone, L.; Guarente, L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef]
- Masoro, E.J. Overview of caloric restriction and ageing. Mech. Ageing Dev. 2005, 126, 913–922. [Google Scholar] [CrossRef]
- Most, J.; Tosti, V.; Redman, L.M.; Fontana, L. Calorie restriction in humans: An update. Ageing Res. Rev. 2017, 39, 36–45. [Google Scholar] [CrossRef]
- Hubbard, B.P.; Sinclair, D.A. Small molecule SIRT1 activators for the treatment of aging and age-related diseases. Trends Pharmacol. Sci. 2014, 35, 146–154. [Google Scholar] [CrossRef] [Green Version]
- Howitz, K.T.; Bitterman, K.J.; Cohen, H.Y.; Lamming, D.W.; Lavu, S.; Wood, J.G.; Zipkin, R.E.; Chung, P.; Kisielewski, A.; Zhang, L.-L.; et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 2003, 425, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Adamczewski, J.P.; Seroz, T.; Vermeulen, W.; Tassan, J.P.; Schaeffer, L.; Nigg, E.A.; Hoeijmakers, J.H.; Egly, J.M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 1994, 79, 1093–1101. [Google Scholar] [CrossRef] [Green Version]
- Yee, A.; Nichols, M.A.; Wu, L.; Hall, F.L.; Kobayashi, R.; Xiong, Y. Molecular cloning of CDK7-associated human MAT1, a cyclin-dependent kinase-activating kinase (CAK) assembly factor. Cancer Res. 1995, 55, 6058–6062. [Google Scholar] [PubMed]
- Kelso, T.W.R.; Baumgart, K.; Eickhoff, J.; Albert, T.; Antrecht, C.; Lemcke, S.; Klebl, B.; Meisterernst, M. Cyclin-dependent kinase 7 controls mRNA synthesis by affecting stability of preinitiation complexes, leading to altered gene expression, cell cycle progression, and survival of tumor cells. Mol. Cell. Biol. 2014, 34, 3675–3688. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, T.; Kwiatkowski, N.; Abraham, B.J.; Lee, T.I.; Xie, S.; Yuzugullu, H.; Von, T.; Li, H.; Lin, Z.; et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 2015, 163, 174–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayrol, F.; Praditsuktavorn, P.; Fernando, T.M.; Kwiatkowski, N.; Marullo, R.; Calvo-Vidal, M.N.; Phillip, J.; Pera, B.; Yang, S.N.; Takpradit, K.; et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat. Commun. 2017, 8, 14290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenall, S.A.; Lim, Y.C.; Mitchell, C.B.; Ensbey, K.S.; Stringer, B.W.; Wilding, A.L.; O’Neill, G.M.; McDonald, K.L.; Gough, D.J.; Day, B.W.; et al. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis 2017, 6, e336. [Google Scholar] [CrossRef] [PubMed]
- Adams, J. The proteasome: Structure, function, and role in the cell. Cancer Treat. Rev. 2003, 29, 3–9. [Google Scholar] [CrossRef]
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. et Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Manasanch, E.E.; Orlowski, R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017, 14, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Nygaard, H.B.; Van Dyck, C.H.; Strittmatter, S.M. Fyn kinase inhibition as a novel therapy for Alzheimer’s disease. Alzheimers Res. 2014, 6, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, A.C.; Salazar, S.V.; Haas, L.T.; Yang, J.; Kostylev, M.A.; Jeng, A.T.; Robinson, S.A.; Gunther, E.C.; Van, C.D.; Nygaard, H.B.; et al. Fyn inhibition rescues established memory and synapse loss in Alzheimer mice. Ann. Neurol. 2015, 77, 953–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ittner, L.M.; Ke, Y.D.; Delerue, F.; Bi, M.; Gladbach, A.; Van Eersel, J.; Wölfing, H.; Chieng, B.C.; Christie, M.J.; Napier, I.A.; et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 2010, 142, 387–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Um, J.W.; Nygaard, H.B.; Heiss, J.K.; Kostylev, M.A.; Stagi, M.; Vortmeyer, A.; Wisniewski, T.; Gunther, E.C.; Strittmatter, S.M. Alzheimer amyloid-β oligomer bound to postsynaptic prion protein activates Fyn to impair neurons. Nat. Neurosci. 2012, 15, 1227–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argyropoulou, A.; Aligiannis, N.; Trougakos, I.P.; Skaltsounis, A.-L. Natural compounds with anti-ageing activity. Nat. Prod. Rep. 2013, 30, 1412–1437. [Google Scholar] [CrossRef]
- Campisi, J.; Kapahi, P.; Lithgow, G.J.; Melov, S.; Newman, J.C.; Verdin, E. From discoveries in ageing research to therapeutics for healthy ageing. Nature 2019, 571, 183. [Google Scholar] [CrossRef] [Green Version]
- Fibrich, B.D.; Lall, N. Chapter 3-Fighting the Inevitable: Skin Aging and Plants. In Medicinal Plants for Holistic Health and Well-Being; Lall, N., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 77–115. ISBN 978-0-12-812475-8. [Google Scholar]
- Parvez, S.; Kang, M.; Chung, H.-S.; Bae, H. Naturally occurring tyrosinase inhibitors: Mechanism and applications in skin health, cosmetics and agriculture industries. Phytother. Res. 2007, 21, 805–816. [Google Scholar] [CrossRef]
- Kusumawati, I.; Indrayanto, G. Chapter 15—Natural Antioxidants in Cosmetics. In Studies in Natural Products Chemistry; Atta-ur-Rahman, Ed.; Elsevier: Amsterdam, The Netherlands, 2013; Volume 40, pp. 485–505. [Google Scholar]
- Martins, A.; Vieira, H.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef] [Green Version]
- Corinaldesi, C.; Barone, G.; Marcellini, F.; Dell’Anno, A.; Danovaro, R. Marine Microbial-Derived Molecules and Their Potential Use in Cosmeceutical and Cosmetic Products. Mar. Drugs 2017, 15, 118. [Google Scholar] [CrossRef]
- Parvez, S.; Kang, M.; Chung, H.-S.; Cho, C.; Hong, M.-C.; Shin, M.-K.; Bae, H. Survey and mechanism of skin depigmenting and lightening agents. Phytother. Res. 2006, 20, 921–934. [Google Scholar] [CrossRef]
- Skropeta, D.; Pastro, N.; Zivanovic, A. Kinase Inhibitors from Marine Sponges. Mar. Drugs 2011, 9, 2131–2154. [Google Scholar] [CrossRef] [Green Version]
- Ning, C.; Wang, H.-M.D.; Gao, R.; Chang, Y.-C.; Hu, F.; Meng, X.; Huang, S.-Y. Marine-derived protein kinase inhibitors for neuroinflammatory diseases. Biomed. Eng. Online 2018, 17, 46. [Google Scholar] [CrossRef]
- Li, T.; Wang, N.; Zhang, T.; Zhang, B.; Sajeevan, T.P.; Joseph, V.; Armstrong, L.; He, S.; Yan, X.; Naman, C.B. A Systematic Review of Recently Reported Marine Derived Natural Product Kinase Inhibitors. Mar. Drugs 2019, 17, 493. [Google Scholar] [CrossRef] [Green Version]
- Della Sala, G.; Agriesti, F.; Mazzoccoli, C.; Tataranni, T.; Costantino, V.; Piccoli, C. Clogging the Ubiquitin-Proteasome Machinery with Marine Natural Products: Last Decade Update. Mar. Drugs 2018, 16, 467. [Google Scholar] [CrossRef] [Green Version]
- Fenical, W.; Jensen, P.R.; Palladino, M.A.; Lam, K.S.; Lloyd, G.K.; Potts, B.C. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 2009, 17, 2175–2180. [Google Scholar] [CrossRef] [Green Version]
- Pettit, G.R.; Tan, R.; Gao, F.; Williams, M.D.; Doubek, D.L.; Boyd, M.R.; Schmidt, J.M.; Chapuis, J.C.; Hamel, E. Isolation and structure of halistatin 1 from the eastern Indian Ocean marine sponge Phakellia carteri. J. Org. Chem. 1993, 58, 2538–2543. [Google Scholar] [CrossRef]
- Pettit, G.R.; Gao, F.; Doubek, D.L.; Boyd, M.R.; Hamel, E.; Bai, R.; Schmidt, J.M.; Tackett, L.P.; Ruetzler, K. Antineoplastic Agents. Part 252. Isolation and Structure of Halistatin 2 from the Comoros Marine Sponge Axinella carteri. Gazz. Chim. Ital. 1993, 123, 371–377. [Google Scholar] [CrossRef]
- Pettit, G.R.; Tan, R.; Herald, D.L.; Williams, M.D.; Cerny, R.L. Antineoplastic Agents. 277. Isolation and Structure of Phakellistatin 3 and Isophakellistatin 3 from a Republic of Comoros Marine Sponge. J. Org. Chem. 1994, 59, 1593–1595. [Google Scholar] [CrossRef]
- Pettit, G.R.; Gao, F.; Schmidt, J.M.; Chapuis, J.-C.; Cerny, R.L. Isolation and structure of axinastatin 5 from a republic of comoros marine sponge. Bioorg. Med. Chem. Lett. 1994, 4, 2935–2940. [Google Scholar] [CrossRef]
- Rudi, A.; Aknin, M.; Gaydou, E.M.; Kashman, Y.; Sodwanones, K.L.M. New triterpenes from the marine sponge Axinella weltneri. J. Nat. Prod. 1997, 60, 700–703. [Google Scholar] [CrossRef]
- Gauvin, A.; Smadja, J.; Aknin, M.; Faure, R.; Gaydou, E.-M. Isolation of bioactive 5α,8α-epidioxy sterols from the marine sponge Luffariella cf. variabilis. Can. J. Chem. 2000, 78, 986–992. [Google Scholar] [CrossRef]
- Campos, P.-E.; Pichon, E.; Moriou, C.; Clerc, P.; Trépos, R.; Frederich, M.; De Voogd, N.; Hellio, C.; Gauvin-Bialecki, A.; Al-Mourabit, A. New Antimalarial and Antimicrobial Tryptamine Derivatives from the Marine Sponge Fascaplysinopsis reticulata. Mar. Drugs 2019, 17, 167. [Google Scholar] [CrossRef] [Green Version]
- Campos, P.-E.; Herbette, G.; Chendo, C.; Clerc, P.; Tintillier, F.; De Voogd, N.J.; Papanagnou, E.-D.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; et al. Osirisynes G-I, New Long-Chain Highly Oxygenated Polyacetylenes from the Mayotte Marine Sponge Haliclona sp. Mar. Drugs 2020, 18, 350. [Google Scholar] [CrossRef]
- Harwood, C.R.; Mouillon, J.-M.; Pohl, S.; Arnau, J. Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 2018, 42, 721–738. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine Bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, M.T.H.D.; Thomas, T. Diversity, host-specificity and stability of sponge-associated fungal communities of co-occurring sponges. PeerJ 2018, 6, e4965. [Google Scholar] [CrossRef] [Green Version]
- Naim, M.A.; Smidt, H.; Sipkema, D. Fungi found in Mediterranean and North Sea sponges: How specific are they? PeerJ 2017, 5, e3722. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Liu, F.; Karuppiah, V.; Ren, Y.; Li, Z. Comparisons of the fungal and protistan communities among different marine sponge holobionts by pyrosequencing. Microb. Ecol. 2014, 67, 951–961. [Google Scholar] [CrossRef]
- Gao, Z.; Li, B.; Zheng, C.; Wang, G. Molecular detection of fungal communities in the hawaiian marine sponges Suberites zeteki and Mycale armata. Appl. Environ. Microbiol. 2008, 74, 6091–6101. [Google Scholar] [CrossRef] [Green Version]
- Kubanek, J.; Jensen, P.R.; Keifer, P.A.; Sullards, M.C.; Collins, D.O.; Fenical, W. Seaweed resistance to microbial attack: A targeted chemical defense against marine fungi. Proc. Natl. Acad. Sci. USA 2003, 100, 6916–6921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gloeckner, V.; Wehrl, M.; Moitinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlik, J.R.; Lindquist, N.L.; Erpenbeck, D.; Wörheide, G.; Hentschel, U. The HMA-LMA dichotomy revisited: An electron microscopical survey of 56 sponge species. Biol. Bull. 2014, 227, 78–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helber, S.B.; Steinert, G.; Wu, Y.-C.; Rohde, S.; Hentschel, U.; Muhando, C.A.; Schupp, P.J. Sponges from Zanzibar host diverse prokaryotic communities with potential for natural product synthesis. FEMS Microbiol. Ecol. 2019, 95. [Google Scholar] [CrossRef] [PubMed]
- Voogd, N.J.; De Gauvin-Bialecki, A.; Polónia, A.R.M.; Cleary, D.F.R. Assessing the bacterial communities of sponges inhabiting the remote western Indian Ocean island of Mayotte. Mar. Ecol. 2018, 39, e12517. [Google Scholar] [CrossRef]
- Rua, C.P.J.; Gregoracci, G.B.; Santos, E.O.; Soares, A.C.; Francini-Filho, R.B.; Thompson, F. Potential metabolic strategies of widely distributed holobionts in the oceanic archipelago of St Peter and St Paul (Brazil). FEMS Microbiol. Ecol. 2015, 91, fiv043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteves, A.I.S.; Amer, N.; Nguyen, M.; Thomas, T. Sample processing impacts the viability and cultivability of the sponge microbiome. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Reynolds, D.; Liu, M.; Stark, M.; Kjelleberg, S.; Webster, N.S.; Thomas, T. Functional equivalence and evolutionary convergence in complex communities of microbial sponge symbionts. Proc. Natl. Acad. Sci. USA 2012, 109, E1878–E1887. [Google Scholar] [CrossRef] [Green Version]
- Moitinho-Silva, L.; Steinert, G.; Nielsen, S.; Hardoim, C.C.P.; Wu, Y.-C.; McCormack, G.P.; López-Legentil, S.; Marchant, R.; Webster, N.; Thomas, T.; et al. Predicting the HMA-LMA status in marine sponges by machine learning. Front. Microbiol. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Tabares, P.; Pimentel-Elardo, S.M.; Schirmeister, T.; Hünig, T.; Hentschel, U. Anti-protease and immunomodulatory activities of bacteria associated with caribbean sponges. Mar. Biotechnol. 2011, 13, 883–892. [Google Scholar] [CrossRef]
- Souza, D.T.; Silva, F.S.P.; Da Silva, L.J.; Da Crevelin, E.J.; Moraes, L.A.B.; Zucchi, T.D.; Melo, I.S. Saccharopolyspora spongiae sp. nov., a novel actinomycete isolated from the marine sponge Scopalina ruetzleri (Wiedenmayer, 1977). Int. J. Syst. Evol. Microbiol. 2017, 67, 2019–2025. [Google Scholar] [CrossRef]
- Vicente, J.; Stewart, A.; Song, B.; Hill, R.T.; Wright, J.L. Biodiversity of actinomycetes associated with caribbean sponges and their potential for natural product discovery. Mar. Biotechnol. 2013, 15, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Aalbersberg, W. Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 2013, 97, 9291–9321. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55, 1759–1766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.K.; Garson, M.J.; Fuerst, J.A. Marine actinomycetes related to the “Salinospora” group from the Great Barrier Reef sponge Pseudoceratina clavata. Environ. Microbiol. 2005, 7, 509–518. [Google Scholar] [CrossRef]
- Xie, Q.-Y.; Wang, C.; Wang, R.; Qu, Z.; Lin, H.-P.; Goodfellow, M.; Hong, K. Jishengella endophytica gen. nov., sp. nov., a new member of the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2011, 61, 1153–1159. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhu, H.; Xu, Q.; Lin, H.; Lu, Y. Micromonospora craniellae sp. nov., isolated from a marine sponge, and reclassification of Jishengella endophytica as Micromonospora endophytica comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 715–720. [Google Scholar] [CrossRef]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Vysotskii, M.V.; Svetashev, V.I.; Nedashkovskaya, O.I.; Gorshkova, N.M.; Mikhailov, V.V.; Yumoto, N.; Shigeri, Y.; Taguchi, T.; Yoshikawa, S. Characterization of Bacillus strains of marine origin. Int. Microbiol. 1999, 2, 267–271. [Google Scholar]
- Bibi, F.; Yasir, M.; Al-Sofyani, A.; Naseer, M.I.; Azhar, E.I. Antimicrobial activity of bacteria from marine sponge Suberea mollis and bioactive metabolites of Vibrio sp. EA348. Saudi J. Biol. Sci. 2020, 27, 1139–1147. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kwon, S.-W.; Rooney, A.P.; Kim, S.-J. Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int. J. Syst. Evol. Microbiol. 2015, 65, 3487–3492. [Google Scholar] [CrossRef] [PubMed]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [Green Version]
- Román-Ponce, B.; Millán-Aguiñaga, N.; Guillen-Matus, D.; Chase, A.B.; Ginigini, J.G.M.; Soapi, K.; Feussner, K.D.; Jensen, P.R.; Trujillo, M.E. Six novel species of the obligate marine actinobacterium Salinispora, Salinispora cortesiana sp. nov., Salinispora fenicalii sp. nov., Salinispora goodfellowii sp. nov., Salinispora mooreana sp. nov., Salinispora oceanensis sp. nov. and Salinispora vitiensis sp. nov., and emended description of the genus Salinispora. Int. J. Syst. Evol. Microbiol. 2020. [Google Scholar] [CrossRef]
- Bolaños, J.; León, L.F.D.; Ochoa, E.; Darias, J.; Raja, H.A.; Shearer, C.A.; Miller, A.N.; Vanderheyden, P.; Porras-Alfaro, A.; Caballero-George, C. Phylogenetic Diversity of Sponge-Associated Fungi from the Caribbean and the Pacific of Panama and Their In Vitro Effect on Angiotensin and Endothelin Receptors. Mar. Biotechnol. 2015, 17, 533–564. [Google Scholar] [CrossRef]
- Höller, U.; Wright, A.D.; Matthee, G.F.; Konig, G.M.; Draeger, S.; Aust, H.-J.; Schulz, B. Fungi from marine sponges: Diversity, biological activity and secondary metabolites. Mycol. Res. 2000, 104, 1354–1365. [Google Scholar] [CrossRef]
- Wang, G.; Li, Q.; Zhu, P. Phylogenetic diversity of culturable fungi associated with the hawaiian sponges Suberites zeteki and Gelliodes fibrosa. Antonie Van Leeuwenhoek 2008, 93, 163–174. [Google Scholar] [CrossRef]
- Ding, B.; Yin, Y.; Zhang, F.; Li, Z. Recovery and phylogenetic diversity of culturable fungi associated with marine sponges Clathrina luteoculcitella and Holoxea sp. in the South China Sea. Mar. Biotechnol. 2011, 13, 713–721. [Google Scholar] [CrossRef]
- Wang, M.; Liu, F.; Crous, P.W.; Cai, L. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 2017, 39, 118–142. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef]
- Jensen, P.R.; Moore, B.S.; Fenical, W. The marine actinomycete genus Salinispora: A model organism for secondary metabolite discovery. Nat. Prod. Rep. 2015, 32, 738–751. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Liu, F.; Liu, S.; Li, H.; Ling, P.; Zhu, X. Poly-γ-glutamate from Bacillus subtilis inhibits tyrosinase activity and melanogenesis. Appl. Microbiol. Biotechnol. 2013, 97, 9801–9809. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Leutou, A.S.; Jeong, H.; Kim, D.; Seong, C.N.; Nam, S.-J.; Lim, K.-M. Anti-Pigmentary Effect of (-)-4-Hydroxysattabacin from the Marine-Derived Bacterium Bacillus sp. Mar. Drugs 2017, 15, 138. [Google Scholar] [CrossRef] [Green Version]
- Dey, G.; Bharti, R.; Dhanarajan, G.; Das, S.; Dey, K.K.; Kumar, B.N.P.; Sen, R.; Mandal, M. Marine lipopeptide Iturin A inhibits Akt mediated GSK3β and FoxO3a signaling and triggers apoptosis in breast cancer. Sci. Rep. 2015, 5, 10316. [Google Scholar] [CrossRef] [Green Version]
- Niggemann, J.; Bozko, P.; Bruns, N.; Wodtke, A.; Gieseler, M.T.; Thomas, K.; Jahns, C.; Nimtz, M.; Reupke, I.; Brüser, T.; et al. Baceridin, a cyclic hexapeptide from an epiphytic Bacillus strain, inhibits the proteasome. ChemBioChem 2014, 15, 1021–1029. [Google Scholar] [CrossRef]
- Candela, T.; Fouet, A. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 2006, 60, 1091–1098. [Google Scholar] [CrossRef]
- Siddiquee, S. Chapter 4—Recent Advancements on the Role of Biologically Active Secondary Metabolites from Aspergillus. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Rodriguez-Couto, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 69–94. ISBN 978-0-444-63501-3. [Google Scholar]
- El-Kady, I.A.; Zohri, A.N.A.; Hamed, S.R. Kojic Acid Production from Agro-Industrial By-Products Using Fungi. Biotechnol. Res. Int. 2014, 2014, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Lateff, A. Chaetominedione, a new tyrosine kinase inhibitor isolated from the algicolous marine fungus Chaetomium sp. Tetrahedron Lett. 2008, 49, 6398–6400. [Google Scholar] [CrossRef]
- Woodhouse, J.N.; Fan, L.; Brown, M.V.; Thomas, T.; Neilan, B.A. Deep sequencing of non-ribosomal peptide synthetases and polyketide synthases from the microbiomes of Australian marine sponges. ISME J. 2013, 7, 1842–1851. [Google Scholar] [CrossRef] [Green Version]
- Vicente, J.; Stewart, A.K.; Van Wagoner, R.M.; Elliott, E.; Bourdelais, A.J.; Wright, J.L.C. Monacyclinones, new angucyclinone metabolites isolated from Streptomyces sp. M7_15 associated with the puerto rican sponge Scopalina ruetzleri. Mar. Drugs 2015, 13, 4682–4700. [Google Scholar] [CrossRef] [Green Version]
- Bode, H.B.; Bethe, B.; Höfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Kjer, J.; Debbab, A.; Aly, A.H.; Proksch, P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010, 5, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Simister, R.L.; Schmitt, S.; Taylor, M.W. Evaluating methods for the preservation and extraction of DNA and RNA for analysis of microbial communities in marine sponges. J. Exp. Mar. Biol. Ecol. 2011, 397, 38–43. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Lee, J.C.; Park, H.R.; Park, D.J.; Lee, H.B.; Kim, Y.B.; Kim, C.J. Improved production of teicoplanin using adsorbent resin in fermentations. Lett. Appl. Microbiol. 2003, 37, 196–200. [Google Scholar] [CrossRef]
- Goff, G.L.; Adelin, E.; Cortial, S.; Servy, C.; Ouazzani, J. Application of solid-phase extraction to agar-supported fermentation. Bioprocess Biosyst Eng 2013, 36, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Angelis, A.; Aligiannis, N.; Rosalia, M.; Abedini, A.; Bakiri, A.; Reynaud, R.; Nuzillard, J.-M.; Gangloff, S.C.; Skaltsounis, A.-L.; et al. In vitro dermo-cosmetic evaluation of bark extracts from common temperate trees. Planta Med. 2016, 82, 1351–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Yamashita, D.; Takeda, Y.; Yonemori, S. Screening for tyrosinase inhibitors among extracts of seashore plants and identification of potent inhibitors from Garcinia subelliptica. Biosci. Biotechnol. Biochem. 2005, 69, 197–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Targeted Region | No of Reads (SI 97%) | No. of Quality-Filtered Reads | No. of OTUs | Richness | Diversity | |
|---|---|---|---|---|---|---|
| Observed OTUs | Chao1 | Shannon | ||||
| V1–V3 | 20806 | 16353 | 337 | 286.74 ± 98.97 | 308.97 ± 94.37 | 5.65 ± 0.87 |
| V3–V4 | 49235 | 47630 | 408 | 315.79 ± 119.56 | 361.43 ± 118.27 | 4.04 ± 0.69 |
| V4–V5 | 39094 | 37213 | 355 | 313.23 ± 103.65 | 324.04 ± 101.00 | 5.39 ± 0.82 |
| ITS2 | 46442 | 46370 | 32 | 26.88 ± 9.18 | 28.92 ± 9.21 | 0.52 ± 0.02 |
| Extracts | Dose-Response Inhibition | ||
|---|---|---|---|
| 0.033 µg/mL | 0.0033 µg/mL | 0.00033 µg/mL | |
| SH-45-EA-SM | Yes | Yes | No |
| SH-54-EA-SM | Yes | Yes | No |
| SH-78-EA-SM | Yes | Yes | No |
| Extracts | Dose-Response Inhibition | ||
|---|---|---|---|
| 0.033 µg/mL | 0.0033 µg/mL | 0.00033 µg/mL | |
| SH-45-EA-SM | Yes | Yes | No |
| SH-54-EA-SM | Yes | Yes | No |
| SH-78-EA-SM | Yes | Yes | Yes |
| SH-115-EA-SM | Yes | Yes | No |
| SH-04-EA-SM | Yes | Yes | Yes |
| SH-68a-EA-LM | Yes | Yes | No |
| SH-68b-EA-LM | Yes | Yes | No |
| SH-116a-EA-SM | Yes | Yes | No |
| SH-123-MeOH-LM | Yes | Yes | No |
| SH-123-MeOH-SM | Yes | Yes | No |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Said Hassane, C.; Fouillaud, M.; Le Goff, G.; Sklirou, A.D.; Boyer, J.B.; Trougakos, I.P.; Jerabek, M.; Bignon, J.; de Voogd, N.J.; Ouazzani, J.; et al. Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process. Microorganisms 2020, 8, 1262. https://doi.org/10.3390/microorganisms8091262
Said Hassane C, Fouillaud M, Le Goff G, Sklirou AD, Boyer JB, Trougakos IP, Jerabek M, Bignon J, de Voogd NJ, Ouazzani J, et al. Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process. Microorganisms. 2020; 8(9):1262. https://doi.org/10.3390/microorganisms8091262
Chicago/Turabian StyleSaid Hassane, Charifat, Mireille Fouillaud, Géraldine Le Goff, Aimilia D. Sklirou, Jean Bernard Boyer, Ioannis P. Trougakos, Moran Jerabek, Jérôme Bignon, Nicole J. de Voogd, Jamal Ouazzani, and et al. 2020. "Microorganisms Associated with the Marine Sponge Scopalina hapalia: A Reservoir of Bioactive Molecules to Slow Down the Aging Process" Microorganisms 8, no. 9: 1262. https://doi.org/10.3390/microorganisms8091262